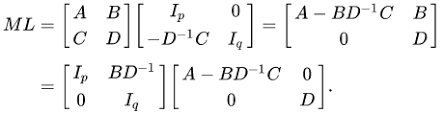Brief introduction to Schur Complement.
This is just a brief introduction of the properties of Schur Complement. Schur Complement has many applications in numerical analysis, such as optimization, machine learning algorithms, or probability and matrix theories.
Let’s begin with the definition of the Schur Complement.
$\large{Definition}$:
The Schur Complement of a black matrix is defined as;
Let $M$ be $n \times n$ matrix
$$M = \begin{bmatrix} A & B \\ C & D \end{bmatrix}$$
Then, the Schur Complement of the block $D$ of the matrix $M$ is
$$M/D := A - BD^{-1}C$$
if $D$ is invertible.
The Schur Complement of the block $A$ of the matrix $M$ is then,
$$M/A := D - CA^{-1}B$$
if $A$ is invertible.
$$\large{Properties}$$
Let
$A$ is a $p \times p$ matrix,
$D$ is a $q \times q$ matrix, with $n = p + q$
So, $B$ is a $p \times q$ matrix.
Let $M$ be defined as,
$$M = \begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix}$$
Suppose we have a linear system as,
$$Ax + By = c \\ B^{T}x + Dy = d$$
Then,
$$\begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix} \begin{bmatrix} x \\ y \end{bmatrix} = \begin{bmatrix} c \\ d \end{bmatrix}$$
Assuming $D$ is invertible,
then we have $y$ as
$$y = D^{-1}(d - B^{T}x)$$
Then, from the equation,
$$Ax + By = c \\ Ax + B(D^{-1}(d - B^{T}x)) = c$$
Then, we see that is,
$$(A - BD^{-1}B^{T})x = c - BD^{-1}d$$
Here, the $A - BD^{-1}B^{T}$ is the Schur Complement of a block $D$ of matrix $M$.
It follows,
$$x = (A-BD^{-1}B^{T})^{-1}(c-BD^{-1}d) \\ y = D^{-1}(d-B^{T}(A-BD^{-1}B^{T})^{-1}(c-BD^{-1}d))$$
And,
$$x = (A-BD^{-1}B^{T})^{-1}c - (A-BD^{-1}B^{T})^{-1}BD^{-1}d \\ y = -D^{-1}B^{T}(A-BD^{-1}B^{T})^{-1}c + (D^{-1}+D^{-1}B^{T}(A-BD^{-1}B^{T})^{-1}BD^{-1})d$$
The $x$ and $y$ are formed as linear functions associated with $c$ and $d$,
and it can be a formula for an inverse of $M$ in terms of the Schur Complement of $D$ in $M$.
$$\begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix} ^{-1} = \begin{bmatrix} (A-BD^{-1}B^{T})^{-1} & -(A-BD^{-1}B^{T})^{-1}BD^{-1} \\ -D^{-1}B^{T}(A-BD^{-1}B^{T})^{-1} & D^{-1}+D^{-1}B^{T}(A-BD^{-1}B^{T})^{-1}BD^{-1} \end{bmatrix}$$
This can be written as,
$$\begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix} ^{-1} = \begin{bmatrix} (A-BD^{-1}B^{T})^{-1} & 0 \\ -D^{-1}B^{T}(A-BD^{-1}B^{T})^{-1} & D^{-1} \end{bmatrix} \begin{bmatrix} I & -BD^{-1} \\ 0 & I \end{bmatrix}$$
Eventually,
$$\begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix} ^{-1} = \begin{bmatrix} I & 0 \\ -D^{-1}B^{T} & I \end{bmatrix} \begin{bmatrix} (A-BD^{-1}B^{T})^{-1} & 0 \\ 0 & D^{-1} \end{bmatrix} \begin{bmatrix} I & -BD^{-1} \\ 0 & I \end{bmatrix}$$
If we take inverse on both sides,
$$\begin{bmatrix} A & B \\ B^{T} & D \end{bmatrix} = \begin{bmatrix} I & BD^{-1} \\ 0 & I \end{bmatrix} \begin{bmatrix} A-BD^{-1}B^{T} & 0 \\ 0 & D \end{bmatrix} \begin{bmatrix} I & 0 \\ D^{-1}B^{T} & I \end{bmatrix}$$
$$= \begin{bmatrix} I & BD^{-1} \\ 0 & I \end{bmatrix} \begin{bmatrix} A-BD^{-1}B^{T} & 0 \\ 0 & D \end{bmatrix} \begin{bmatrix} I & BD^{-1} \\ 0 & I \end{bmatrix} ^{T}$$
This is done by an assumption with $D$ is invertible, and this decomposition is called Block LDU Decomposition.
Then, we can get another factorization of $M$ if A is invertible.
For any symmetric matrix, $M$,
if D is invertible, then
1) $M \succ 0$ $iff$ $D \succ 0$ and $A-BC^{-1}B^{T} \succ 0$
2) If $C \succ 0$, then $M \succeq 0$ $iff$ $A-BC^{-1}B^{T} \succeq 0$
We can easily get the matrix $M$ in the linear combination and the inverse of the $M$ with Schur Complement.
We also can check if the $M$ is positive semi-definite.
Reference:
Schur Complement
The Schur Complement and Symmetric Positive Semidefinite (and Definite) Matrices

![[hist]](/2019/09/09/hist/histrogram-1.png)
![[hist]](/2019/09/09/hist/histrogram-2.png)
![[hist]](/2019/09/09/hist/histrogram-3.png)
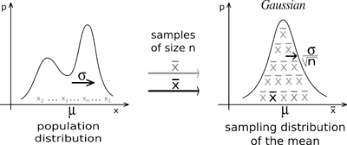
![[hist]](/2019/09/09/CLT/CLT-1.png)
![[hist]](/2019/09/09/CLT/CLT-3.png)
![[hist]](/2019/09/09/CLT/CLT-4.png)
![[hist]](/2019/09/09/CLT/CLT-14.png)
![[hist]](/2019/09/09/CLT/CLT-15.png)
![[hist]](/2019/09/09/CLT/CLT-16.png)